# Load required libraries
library(randomForest)
Type rfNews() to see new features/changes/bug fixes.
Attaching package: 'ggplot2'
The following object is masked from 'package:randomForest':
margin
# Set seed for reproducibility
set.seed(123)
data <- mtcars
Split data into training and testing sets
train_indices <- sample(1:nrow(data), 0.7 * nrow(data))
train_data <- data[train_indices, ]
test_data <- data[-train_indices, ]
Train Random Forest model and make predictions
rf_model <- randomForest(mpg ~ ., data = train_data, ntree = 500)
predictions <- predict(rf_model, newdata = test_data)
Calculate RMSE
rmse <- sqrt(mean((test_data$mpg - predictions)^2))
cat("Root Mean Square Error:", rmse, "\n")
Root Mean Square Error: 2.00452
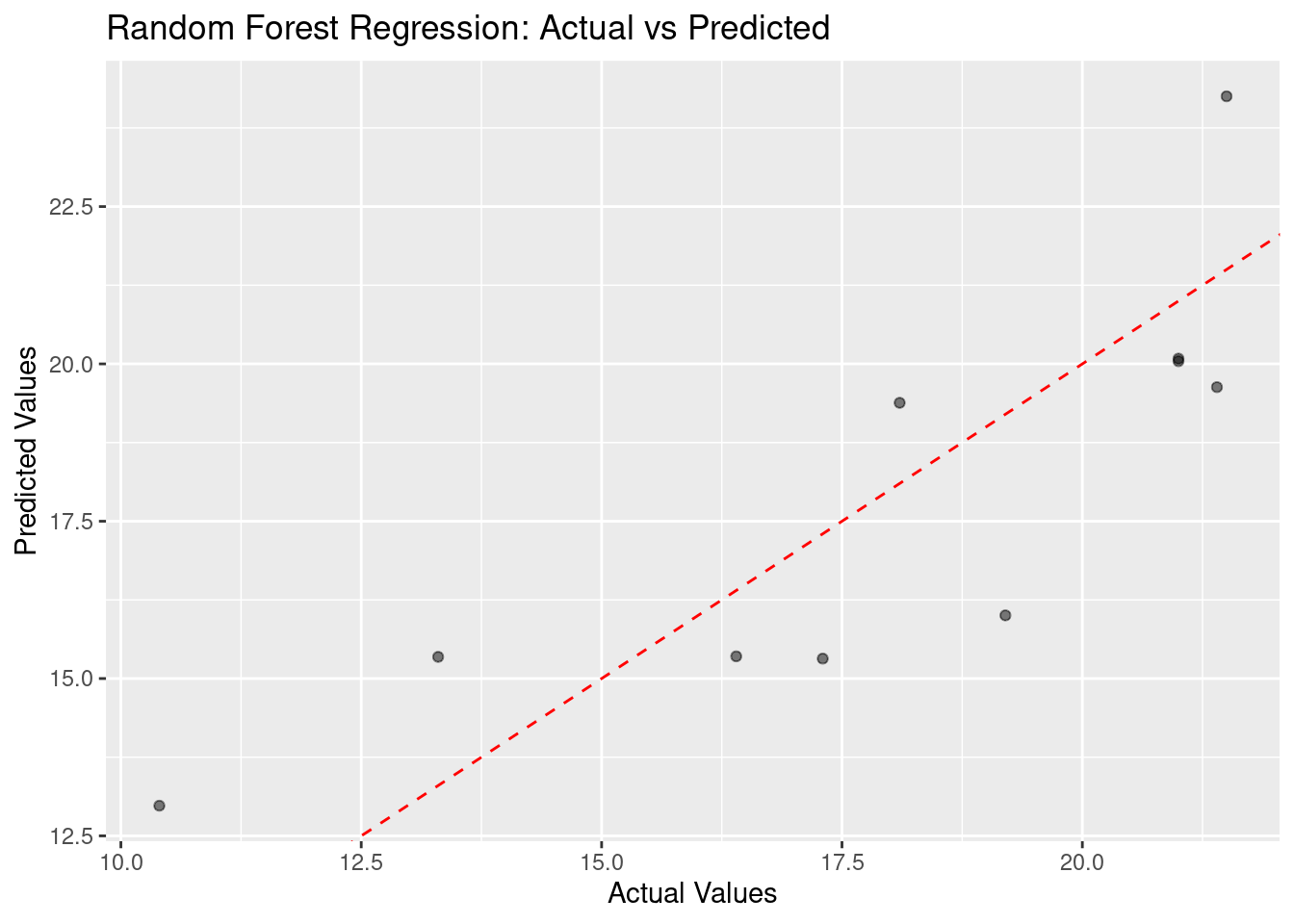
Plot actual vs predicted values
ggplot(data.frame(actual = test_data$mpg, predicted = predictions), aes(x = actual, y = predicted)) +
geom_point(alpha = 0.5) +
geom_abline(intercept = 0, slope = 1, color = "red", linetype = "dashed") +
labs(x = "Actual Values", y = "Predicted Values", title = "Random Forest Regression: Actual vs Predicted")
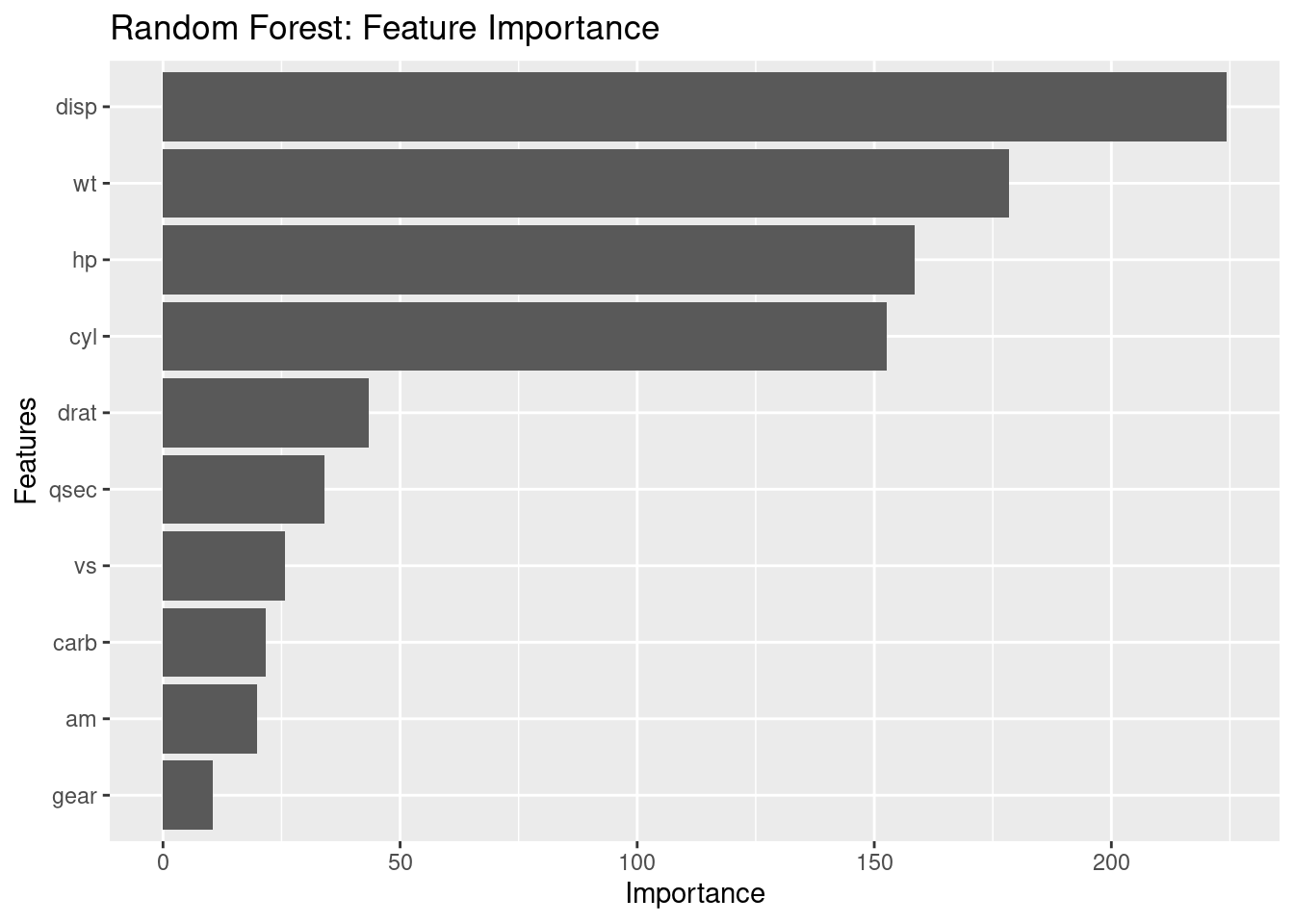
Print feature importance
importance <- importance(rf_model)
print(importance)
IncNodePurity
cyl 152.64821
disp 224.33387
hp 158.42647
drat 43.37372
wt 178.44424
qsec 34.05620
vs 25.67666
am 19.73485
gear 10.54314
carb 21.62506
Plot feature importance
importance_df <- data.frame(feature = rownames(importance), importance = importance[, 1])
ggplot(importance_df, aes(x = reorder(feature, importance), y = importance)) +
geom_bar(stat = "identity") +
coord_flip() +
labs(x = "Features", y = "Importance", title = "Random Forest: Feature Importance")
alternative at https://hackernoon.com/random-forest-regression-in-r-code-and-interpretation