x = 5
y <- 8
z <- x+y
zWeek 10 - Introduction to R
Which programs you should download and install
- R programming language (required) (the link for CRAN project).
- RStudio (Not required but strongly recommended) (the website of RStudio)
What the R part of the course will cover
In the following two weeks you will learn:
- Introduction to R.
- Data types in R.
- How to import data in R.
- Checking summary stats and making exploratory data analysis with R.
- Using
dplyrto manipulate data with R. - Using
ggplotfor data visualization with R.
What is R?
- R is a suite of software facilities for:
- Reading and manipulating data
- Computation
- Conducting statistical data analysis
- Application and development of Machine Learning Algorithms
- Displaying the results
- R is the open-source version (i.e. freely available version - no license fee) of the S programming language, a language for manipulating objects.
- Software and packages can be downloaded from the link for CRAN project

RStudio
- The R Console by itself is not very interesting and useful. A tool named RStudio is designed to use R more efficient and easily.
- RStudio can be installed after installing R.
- RStudio won’t work without R. R has to be installed on your computer.
- You can think RStudio as an upgrade of R, on visual and functionality terms. It doesn’t add anything that R cannot do.
- RStudio can be downloaded from the website of RStudio
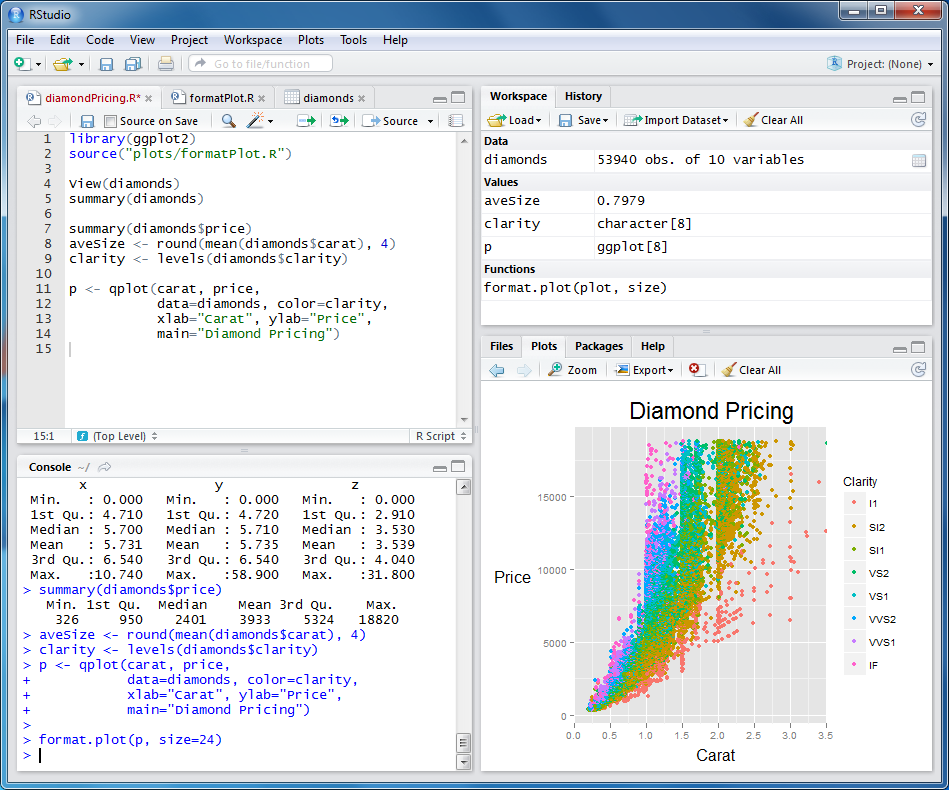
- RStudio have 4 main parts:
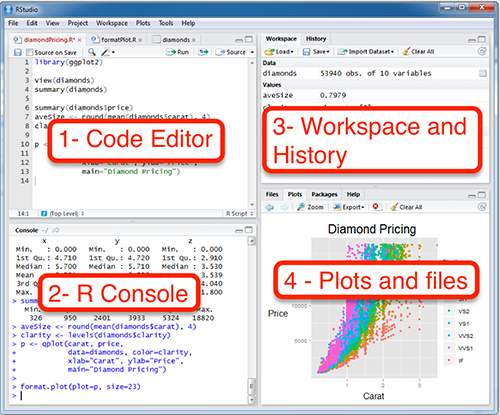
Using R and RStudio from Posit Cloud
If you are a beginner at R and will only use it for the lecture then you may not want to install it into your computer.
A great alternative for this is to use Posit Cloud.
Posit is the company that is the founder of RStudio and provides a cloud solution for using R and Rstudio.
First, go to the website of Posit
Click Sign Up at the upper right part of the page
- Then, select the free plan and click Learn more.
- The Free Plan gives you 25 hours of computing time per month which should be sufficient for this course.
- If you ever think that you are going to exceed the limit you can always download and install R and RStudio to your computer for free.
- Click Sign Up at the next page.
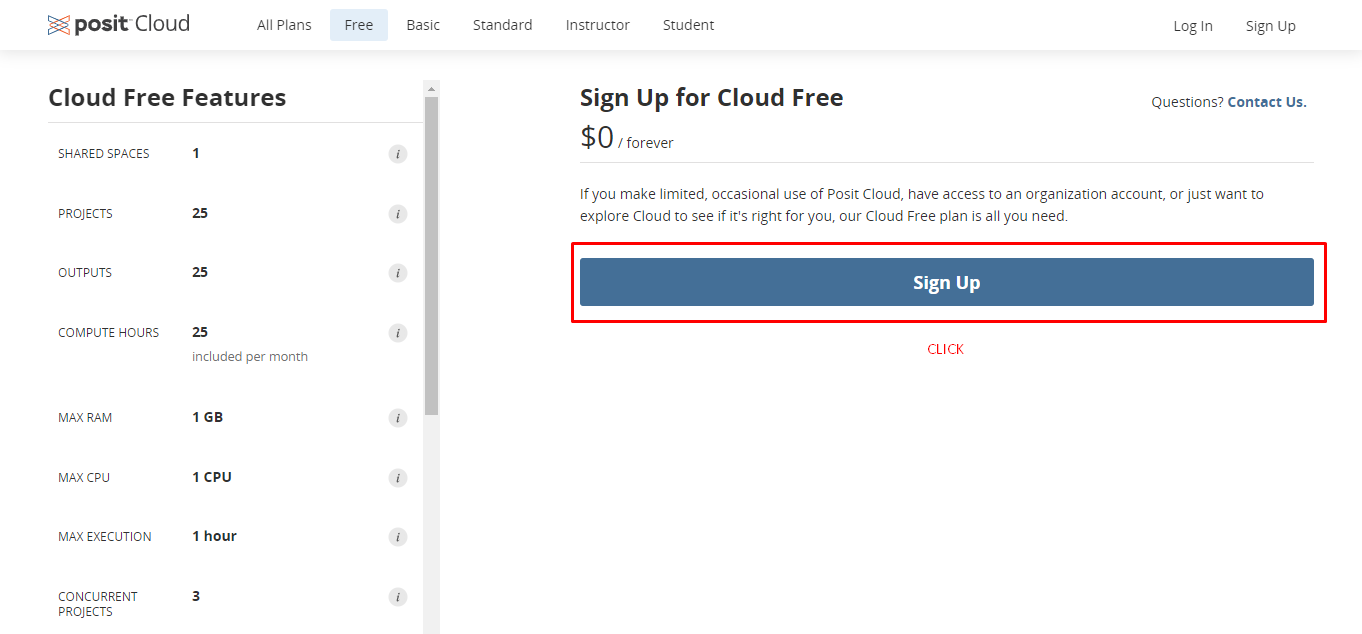
- Enter your e-mail, a password, your name and your surname to register to posit cloud.
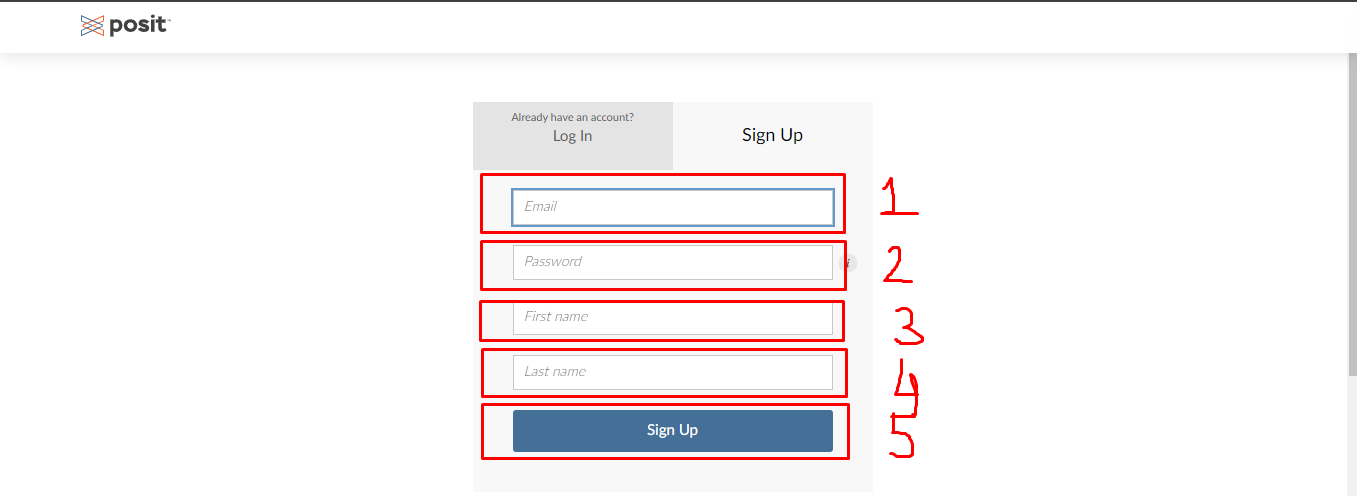
- After registering and possibly confirming your mail address, you can login with your e-mail and password.
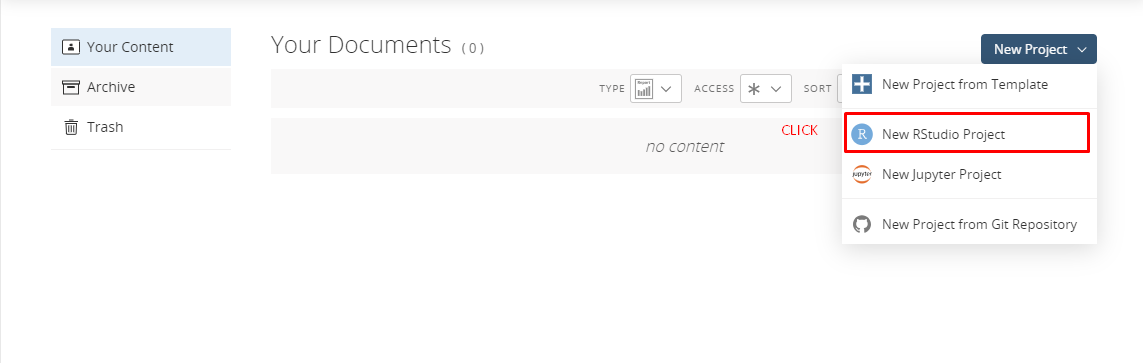
- You will encounter the following page when you successfully login to the Posit Cloud.
- Select New Project and New RStudio Project from the upper right buttons.
- After a short period of deployment time, a fresh Rstudio will open as in the following picture.
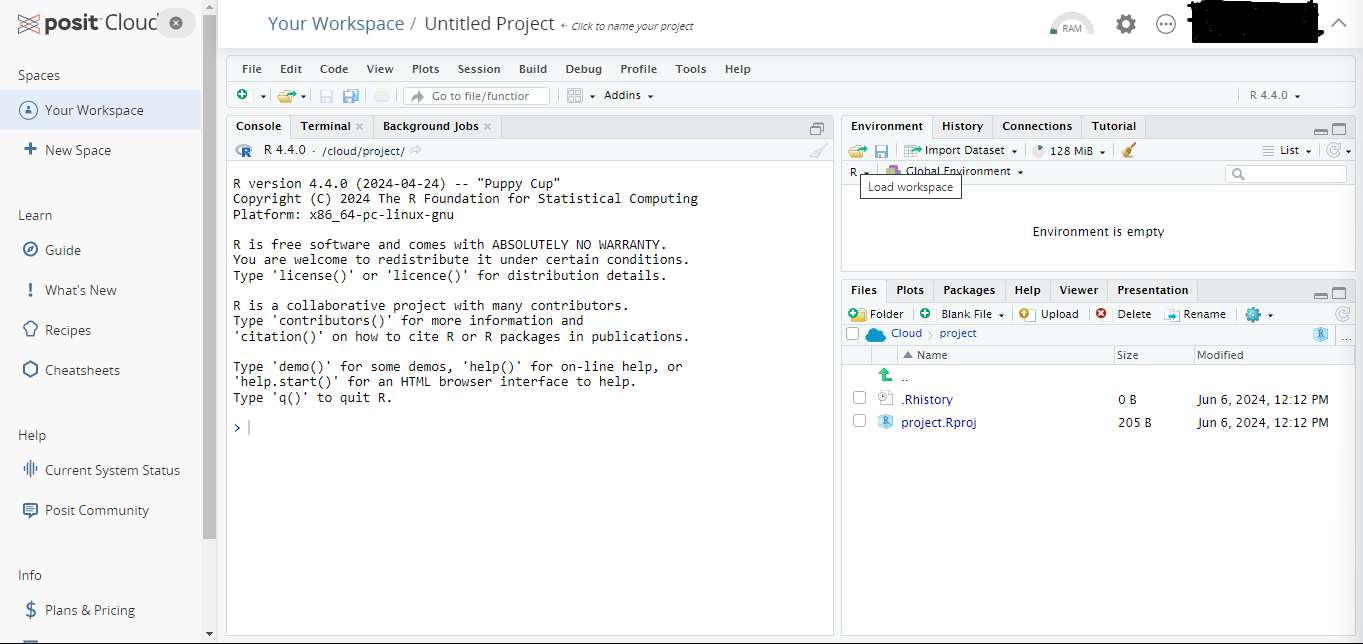
- Give a new name to your project and open a R script to work on from the menu File \(\rightarrow\) New File \(\rightarrow\) R Script.
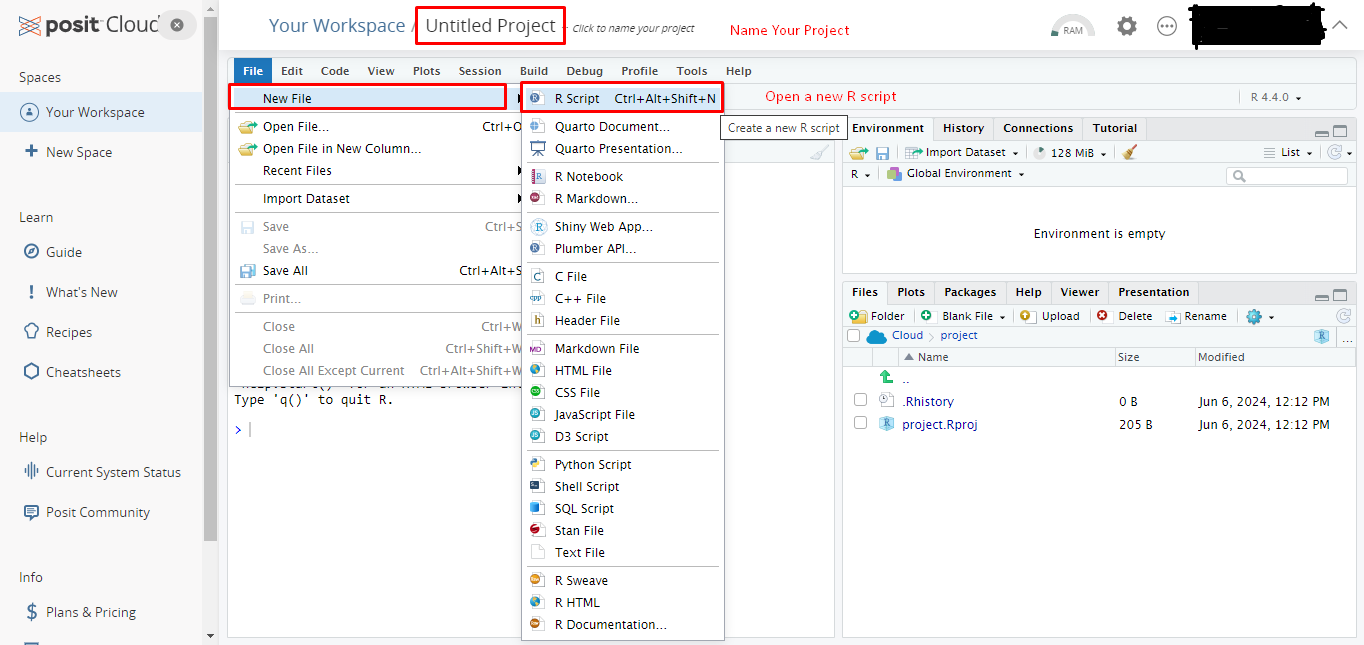
- Now you can start to use RStudio in cloud and make your computations.
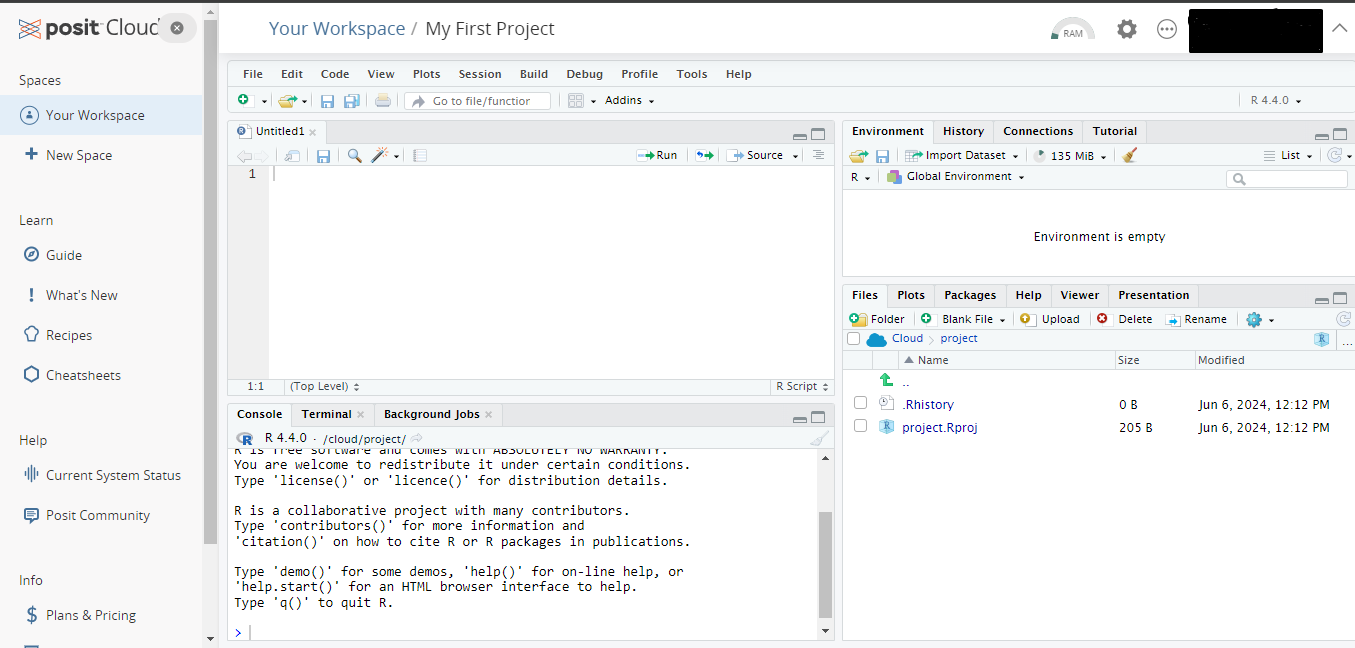
Advantages of R
- Open Source:
- There is no clear difference between user and developer.
- A unique solution for the given problem can be constructed.
- You are not limited to pre-defined options by a fixed user interface as is common in proprietary software.
- Open source also allows to use the program freely without spending any money.
- Flexibility:
- Gives access to the source code, allows to modify and improve it according to the needs.
- Ability to further developments and capacity increase with tools like RStudio and Shiny.
- New packages to solve a certain problem is consistently added to the R repository.
- Ability to produce reports in PDF and HTML format.
- Community:
- R has a lot of material in online platforms, in books and in courses.
- A lot of information can be found via Q&A websites, social media networks, and numerous blogs.
Some useful websites to get help
Typically, a problem you may be encountering is not new and others have faced, solved, and documented the same issue online.
- The following resources can be used to search for online help. Although, I typically just google the problem and find answers relatively quickly.
- RSiteSearch(“key phrase”): searches for the key phrase in help manuals and archived mailing lists on the R Project website at http://search.r-project.org/.
- Stack Overflow: a searchable Q&A site oriented toward programming issues. 75 % of my answers typically come from Stack Overflow questions tagged for R at http://stackoverflow.com/questions/tagged/r.
- Cross Validated: a searchable Q&A site oriented toward statistical analysis. Many questions regarding specific statistical functions in R are tagged for R at http://stats.stackexchange.com/questions/tagged/r.
- R–seek: a Google custom search that is focused on R-specific websites. Located at http://rseek.org/
- R -bloggers: a central hub of content collected from over 500 bloggers who provide news and tutorials about R. Located at http://www.r-bloggers.com/
- ChatGPT obviously.
Basic Calculations and Defining Objects in R
- You can either write a code directly into the console or you can use a script.
- Using a script is more efficient. Because it is easier to write modify and save a R Code in a script.
- Open a script with File \(\rightarrow\) New File \(\rightarrow\) R Script or you can use shortcut Ctrl + Shift + N
Objects
- R works by creating objects and using various functions calls that create and use these objects. For example;
- Vectors of numbers, logical values (TRUE and FALSE), character strings and even complex numbers.
- Matrices and general n-way arrays
- Lists - arbitrary collections of objects of any type; e.g. list of vectors, list of matrices, etc.
- Data frames - a general data set type
- functions (yes even functions are objects)
Defining Variables in R
- R is case sensitive !!!
Basic Math in R
43 + 35 # addition43 - 35 # subtraction12 * 8 # multiplication100 / 8 # division2^4 # power100 %% 8 # remainder100 %/% 8 # dividentLogical Comparisons in R
5 < 82 + 2 == 5T == TRUE3 * 3 == 93 * 3 != 83 * 3 != 9Functions
Functions are special commands that are designed for a particular purpose.
For example
sum()gives the sum of a numerical values,sqrt()takes root of a number etc..Functions are always followed by a
(). Inside the()most of the functions take some special values called arguments.Lets look at the
helppage for thesqrt()function.
?sqrt{r, out.width = "80%", fig.asp=.75, echo=FALSE, fig.align= "center", fig.cap="Help Documentation for sqrt() function"} knitr::include_graphics("./figures/help_sqrt.png")
sqrt()function only takes one argumentxwhich is either a single number, or arrays of numbers.
sqrt(8)sqrt(c(1,4,9,16,25))- 1
- 2
- 3
- 4
- 5
- Lets look at the
helppage for thesum()function.
?sum{r, out.width = "80%", fig.asp=.75, echo=FALSE, fig.align= "center", fig.cap="Help Documentation for sum() function"} knitr::include_graphics("./figures/help_sum.png")
- According to the help file the usage of function is
sum(..., na.rm = FALSE) sum()function takes two arguments....numeric or complex or logical vectors.na.rmlogical. Should missing values (including NaN) be removed?
- The second argument
na.rmhas a default value ofFALSE. A default value means that if you don’t specify a value, it will take the default value, hereFALSE.
x<- c(6, 8, 10, 12, 14)sum(x)sum(x, na.rm = FALSE)sum(x, na.rm = TRUE)y<- c(6, 8, 10, 12, NA)sum(y)sum(y, na.rm = FALSE)sum(y, na.rm = TRUE)z <- c(T, T, F, F, F, T, T)
sum(z)Packages
- Some functions are not contained locally in R. They are called packages and they should be installed when needed.
- R contains one or more libraries of packages. A package contain various functions and data sets for numerous purposes, e.g.
e1071package,spatstatpackage andDandEFApackage, etc. - Some packages are a part of the R. Others should be downloaded from the Comphrensive R Archive Network.
- To access all of the functions and data sets of a particular package; for example, DandEFA; it must be loaded to the workspace:
# install.packages('DandEFA')library(DandEFA) # Buy you have to call and load a package every new R session.- You can also use Rstudio to download multiple packages easily and at once.
Tools -> Install Packages
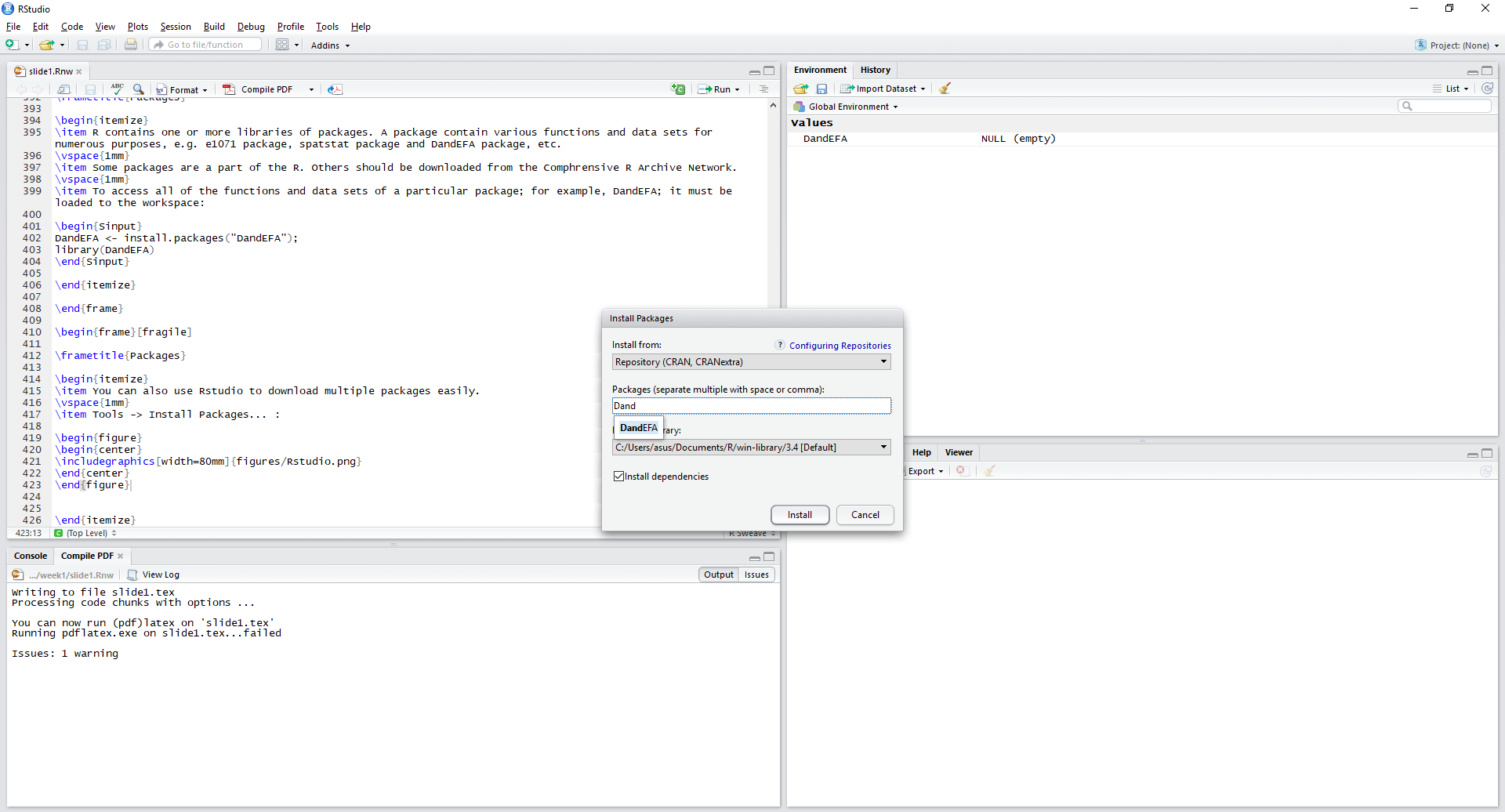
An Example for Packages: DandEFA Package
- Using packages to utilize various methods and algorithms.
- DandEFA package contains functions for a particular analysis called factor analysis.
- Factor Analysis is a method for categorize variables into groups to find the relationship between the variables in the same group.
- The package contains functions:
- factload: A method for producing the factor loadings
- dandelion: A method for visualizing the factor loadings
help(package="DandEFA")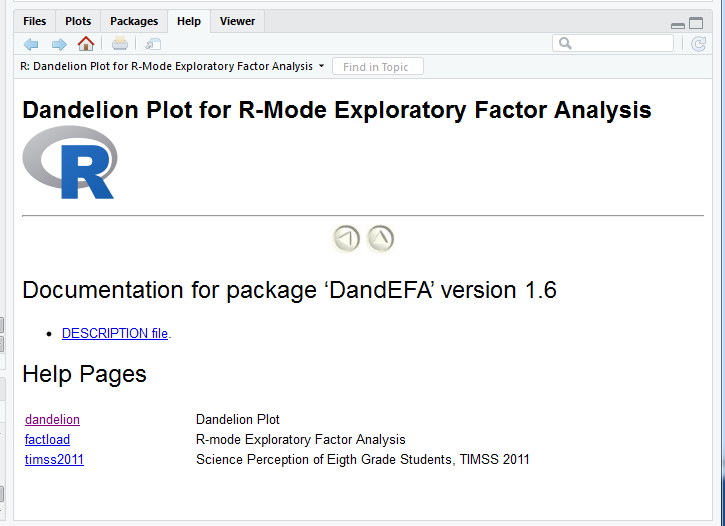
- Alternatively you can also use the bottom-right panel in RStudio to get info on a specific function:
#packageDescription("DandEFA")- You don’t have to understand the following code, but understand that the following code is taken from the documentation from the
DandEFApackage and can be applied directly.
library(DandEFA) # loading the package
data(timss2011) # loading the dataset
timss2011 <- na.omit(timss2011) # removing the rows with missing values
dandpal <- rev(rainbow(100, start = 0, end = 0.2)) # Choose colors for visualisation
facl <- factload(timss2011,nfac=5,method="prax",cormeth="spearman") # Find the factor loadings
facl # Show the factor loadings
dandelion(facl,bound=0,mcex=c(1,1.2),palet=dandpal) # Visualise
Loadings:
[,1] [,2] [,3] [,4] [,5]
X10A 0.103 -0.101 -0.224
X10B
X10C 0.106 -0.129
X11A -0.544 -0.130
X11B -0.514
X11C -0.129 -0.105 -0.500
X11D -0.475
X12A -0.116 -0.152 -0.338 0.318
X12B -0.254 -0.133 -0.328 0.256
X12C -0.149 -0.136 -0.298 0.249
X13A 0.549
X13B 0.504
X13C 0.583
X13D 0.398
X13E 0.595
X13F 0.458
X17A -0.539 -0.419 -0.140
X17B 0.633 0.156 -0.164
X17C -0.350 -0.450 -0.185
X17D 0.727 0.222 -0.173
X17E -0.325 -0.337 -0.164
X17F -0.611 -0.445 -0.143
X17G -0.252 -0.481 -0.145 0.157
X18A -0.303 -0.420 -0.267 -0.138
X18B 0.537 0.146 0.152 -0.152
X18C -0.353 -0.326 -0.192
X18D -0.416 -0.413 -0.277
X18E -0.160 -0.381 -0.239 -0.125
X19A -0.540 -0.443 -0.135 -0.254
X19B 0.633 0.119
X19C 0.694 0.158
X19D -0.519 -0.424 -0.163 -0.256
X19E 0.687 0.112
X19F -0.415 -0.462 -0.124 -0.361
X19G -0.313 -0.491 -0.220 -0.359
X19H -0.383 -0.500 -0.214 -0.361
X19I 0.690
X19J -0.238 -0.507 -0.158
X19K -0.620 0.142
X19L -0.714 0.124
X19M -0.749 0.101
X19N -0.184 -0.654
X21A -0.120 -0.106
[,1] [,2] [,3] [,4] [,5]
SS loadings 5.576 1.851 4.965 1.987 1.076
Proportion Var 0.130 0.043 0.115 0.046 0.025
Cumulative Var 0.130 0.173 0.288 0.334 0.359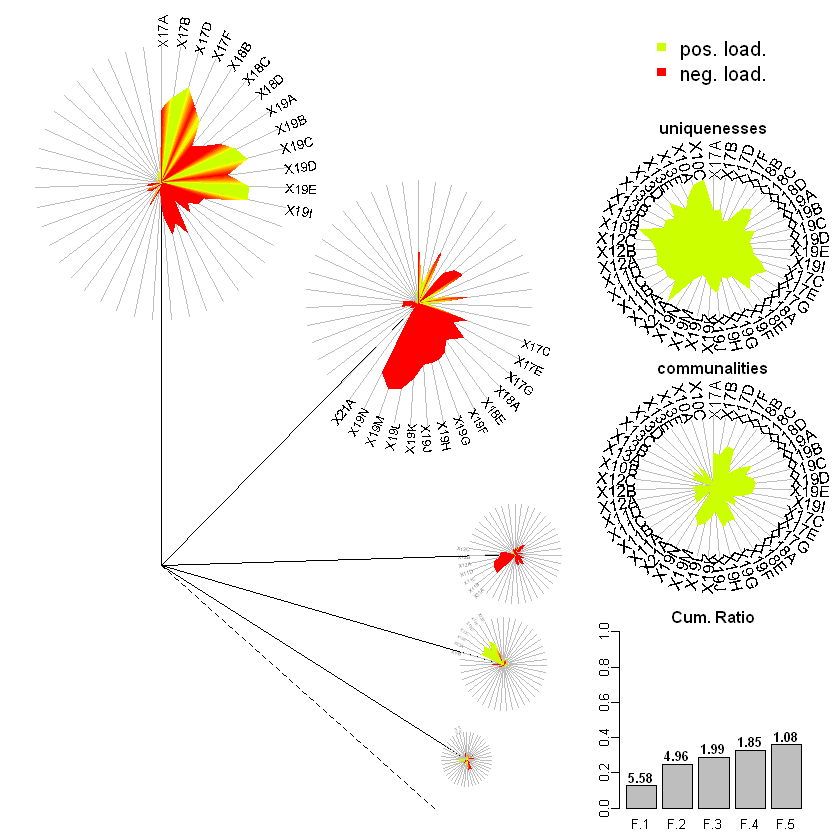
- In summary, packages provide a flexible environment.
- Employing multiple methods and algorithms in the same time
- Programming and using packages are two core elements in R.
Working Directory
- In order to work in R, you should specify a active working directory. In brief this is the location where R will get and save the files.
- You can call the active working directory with the command
getwd() - Note: If you don’t understand the concept of
working directory, you will probably get errors during importing dataset and locating files. So be careful.
# returns path for the current working directory
getwd()- You can change your active working directory either by using
setwd()function or by using Session \(\rightarrow\) Set Working Directory \(\rightarrow\) To Source File Location after saving a script.
# set the working directory to a specified directory
setwd("C:/Users/erhan/Desktop")
getwd()setwd("C:/Users/erhan/Documents/FEF1002")
getwd()Data Types in R Programming
Vectors
- Vectors are one dimensional arrays that keeps only one type of variables.
- All the elements in a vector should be the same type. (Numeric, string, logical etc.)
x <- c(10.4, 5.6, 3.1, 6.4, 21.7) # Numeric Vector
x- 10.4
- 5.6
- 3.1
- 6.4
- 21.7
x <- c("boy","girl","boy","girl","boy","boy") # character vector
x- 'boy'
- 'girl'
- 'boy'
- 'girl'
- 'boy'
- 'boy'
x <- c(TRUE,TRUE,FALSE,TRUE,TRUE,FALSE) # logical vector
x- TRUE
- TRUE
- FALSE
- TRUE
- TRUE
- FALSE
# or you can use
x <- c(T,T,F,T,T,F) # logical vector
x- TRUE
- TRUE
- FALSE
- TRUE
- TRUE
- FALSE
- What if I put different kind of values in a vector.
c(10, 20, 26, T) # numeric and logical values- 10
- 20
- 26
- 1
c(10, 20, 26, "apple") # numeric and string- '10'
- '20'
- '26'
- 'apple'
c(T, F, "apple", "banana") # logical and string- 'TRUE'
- 'FALSE'
- 'apple'
- 'banana'
c(T, "apple", 10) # logical, string, numeric- 'TRUE'
- 'apple'
- '10'
- Accessing elements in a vector is easy,
x <- c(10.4, 5.6, 3.1, 6.4, 21.7) # Numeric VectorIndexing in R starts with
1opposing to some programming languages like Python which starts indexing with0.Select fifth element of the vector.
x[5]- Select first, third and fifth element of the vector.
ind <- c(1,3,5)
x[ind]- 10.4
- 3.1
- 21.7
- Select second and fourth element of the vector.
ind <- c(F,T,F,T,F)
x[ind]- 5.6
- 6.4
- A logical operation over a vector would create a logical vector (important!!)
x <- c(10.4, 5.6, 3.1, 6.4, 21.7) # Numeric Vector- Find whether an element is higher than 7.
ind <- (x > 7)
ind- TRUE
- FALSE
- FALSE
- FALSE
- TRUE
- Find elements that is higher than 7.
x[ind]- 10.4
- 21.7
- Find elements that is equal or lower than 7.
x[!ind]- 5.6
- 3.1
- 6.4
- We will use indices to manipulate data sets later. But a shorter version of the code is
x <- c(10.4, 5.6, 3.1, 6.4, 21.7) # Numeric Vectorx[x > 7]- 10.4
- 21.7
- A logical operator checks whether the both sides have equal length or one side has length 1.
x <- c(10.4, 5.6, 3.1, 6.4, 21.7)
y <- c(4, 7, 8, 2, 35)ind <- (x > y)
ind- TRUE
- FALSE
- FALSE
- TRUE
- FALSE
- If the number of elements are not equal:
x <- c(10.4, 5.6, 3.1, 6.4, 21.7)
y <- c(4,7,8,2)- will give an output but with a warning.
ind <- (x > y)Warning message in x > y:
"longer object length is not a multiple of shorter object length"ind- TRUE
- FALSE
- FALSE
- TRUE
- TRUE
Modifying Vectors
- Any element of the vector can be modified easily:
x <- c(10.4, 5.6, 3.1, 6.4, 21.7)x[4] <- 7.3
x- 10.4
- 5.6
- 3.1
- 7.3
- 21.7
- A group of elements can be modified too
x <- c(10.4, 5.6, 3.1, 6.4, 21.7)x[x > 7] <- 100
x- 100
- 5.6
- 3.1
- 6.4
- 100
- Some advance stuff: (data imputation)
x <- c(10.4, NA, 3.1, 6.4, NA)is.na(x)- FALSE
- TRUE
- FALSE
- FALSE
- TRUE
x[is.na(x)] <- mean(x, na.rm = TRUE)
x- 10.4
- 6.63333333333333
- 3.1
- 6.4
- 6.63333333333333
Manipulating vectors
- Merging vectors with c():
x <- c(10.4, 5.6, 3.1, 6.4, 21.7)
y <- c(4, 7, 8, 2, 35)
z <- c(x,y)z- 10.4
- 5.6
- 3.1
- 6.4
- 21.7
- 4
- 7
- 8
- 2
- 35
- Summation or multiplication over vectors. Note: Again both vectors either have be of same size or one has to be of length one.
x
y- 10.4
- 5.6
- 3.1
- 6.4
- 21.7
- 4
- 7
- 8
- 2
- 35
z <- x + y
z- 14.4
- 12.6
- 11.1
- 8.4
- 56.7
z <- x * y
z- 41.6
- 39.2
- 24.8
- 12.8
- 759.5
Generating Sequences
- the colon
:,
x <- 1:10
x- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
x <- 2*(1:10)
x- 2
- 4
- 6
- 8
- 10
- 12
- 14
- 16
- 18
- 20
- the
seq()function.
x <- seq(1,10)
x- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
x <- seq(1,10,by=0.5)
x- 1
- 1.5
- 2
- 2.5
- 3
- 3.5
- 4
- 4.5
- 5
- 5.5
- 6
- 6.5
- 7
- 7.5
- 8
- 8.5
- 9
- 9.5
- 10
- the
rep()function.
x <- rep(3, 10)
x- 3
- 3
- 3
- 3
- 3
- 3
- 3
- 3
- 3
- 3
y <- rep(c(F,T,F,T,T,T),3)
y- FALSE
- TRUE
- FALSE
- TRUE
- TRUE
- TRUE
- FALSE
- TRUE
- FALSE
- TRUE
- TRUE
- TRUE
- FALSE
- TRUE
- FALSE
- TRUE
- TRUE
- TRUE
z1 <- rep(c(4,7,8,2,35),each=3)
z1- 4
- 4
- 4
- 7
- 7
- 7
- 8
- 8
- 8
- 2
- 2
- 2
- 35
- 35
- 35
z2 <- rep(c(4,7,8,2,35), times = 3)
z2- 4
- 7
- 8
- 2
- 35
- 4
- 7
- 8
- 2
- 35
- 4
- 7
- 8
- 2
- 35
An Example on Vectors
x <- c(2,4,6,8,10)
y <- c("apple", "banana", "peach", "walnut", "apple")sum(x)sum(x < 6)mean(x < 6)x[x < 6]- 2
- 4
x
y- 2
- 4
- 6
- 8
- 10
- 'apple'
- 'banana'
- 'peach'
- 'walnut'
- 'apple'
mean(y=="apple")mean(x > 6 & y=="apple")Factors
A factor is a special type of vector used to represent categorical data, e.g. gender, social class, etc.
- Stored internally as a numeric vector with values \(1, 2, ..., k\), where \(k\) is the number of levels.
- Can have either ordered and unordered factors.
- A factor with \(k\) levels is stored internally consisting of 2 items.
- a vector of \(k\) integers
- a character vector containing strings describing what the \(k\) levels are.
Factor Example
Five people are asked to rate the performance of a product on a scale of 1-5, with 1 representing very poor performance and 5 representing very good performance. The following data were collected.
- We have a numeric vector containing the satisfaction levels.
satisfaction <- c(1, 3, 4, 2, 2, 3, 4, 2, 1, 2, 1, 1, 4, 3)- Want to treat this as a categorical variable and so the second line creates a factor. The
levels=1:5argument indicates that there are 5 levels of the factor. We also set the labels for each factor.
fsatisfaction <- factor(satisfaction,
levels=1:5,
labels = c("very poor", "poor", "average","good", "very good"))fsatisfaction- very poor
- average
- good
- poor
- poor
- average
- good
- poor
- very poor
- poor
- very poor
- very poor
- good
- average
Levels:
- 'very poor'
- 'poor'
- 'average'
- 'good'
- 'very good'
Matrices
- Matrices are used for many purposes in R.
- First let’s create a vector from a normal distribution that we will convert to matrix.
set.seed(100) # to ensure the numbers are same for each of you
m <- rnorm(12,0,1)
m- -0.502192350531457
- 0.131531165327303
- -0.07891708981887
- 0.886784809417845
- 0.116971270510841
- 0.318630087617032
- -0.58179068471591
- 0.714532710891568
- -0.825259425862769
- -0.359862131395465
- 0.0898861437775305
- 0.0962744602851301
dim(m) <- c(3,4)
m| -0.50219235 | 0.8867848 | -0.5817907 | -0.35986213 |
| 0.13153117 | 0.1169713 | 0.7145327 | 0.08988614 |
| -0.07891709 | 0.3186301 | -0.8252594 | 0.09627446 |
- or you can specify the dimensions with the
matrix()function
set.seed(100) # to ensure the numbers are same for each of you
m <- rnorm(12)
m- -0.502192350531457
- 0.131531165327303
- -0.07891708981887
- 0.886784809417845
- 0.116971270510841
- 0.318630087617032
- -0.58179068471591
- 0.714532710891568
- -0.825259425862769
- -0.359862131395465
- 0.0898861437775305
- 0.0962744602851301
m <- matrix(m, nrow = 3, ncol = 4, byrow = F)
m| -0.50219235 | 0.8867848 | -0.5817907 | -0.35986213 |
| 0.13153117 | 0.1169713 | 0.7145327 | 0.08988614 |
| -0.07891709 | 0.3186301 | -0.8252594 | 0.09627446 |
- Basic functions on matrices.
nrow()andncol()calls numbers of rows and columns.t()calls the transpose of the matrix.rownames()andcolnames()are the names of columns and rows.
set.seed(100) # to ensure the numbers are same for each of you
m <- rnorm(12)
m <- matrix(m, nrow = 3, ncol = 4, byrow = F)
m| -0.50219235 | 0.8867848 | -0.5817907 | -0.35986213 |
| 0.13153117 | 0.1169713 | 0.7145327 | 0.08988614 |
| -0.07891709 | 0.3186301 | -0.8252594 | 0.09627446 |
nrow(m)ncol(m)colnames(m) <- c("A", "B", "C", "D")
m| A | B | C | D |
|---|---|---|---|
| -0.50219235 | 0.8867848 | -0.5817907 | -0.35986213 |
| 0.13153117 | 0.1169713 | 0.7145327 | 0.08988614 |
| -0.07891709 | 0.3186301 | -0.8252594 | 0.09627446 |
Merging Vectors
rbind()andcbind()functions merges vectors or matrices into matrices.
set.seed(100)
X1 <- rnorm(12)
X2 <- 1:12m <- cbind(X1,X2)
m| X1 | X2 |
|---|---|
| -0.50219235 | 1 |
| 0.13153117 | 2 |
| -0.07891709 | 3 |
| 0.88678481 | 4 |
| 0.11697127 | 5 |
| 0.31863009 | 6 |
| -0.58179068 | 7 |
| 0.71453271 | 8 |
| -0.82525943 | 9 |
| -0.35986213 | 10 |
| 0.08988614 | 11 |
| 0.09627446 | 12 |
Number of columns should be equal for
rbind.Likewise, number of rows should be equal for
cbind.Create two matrices
set.seed(100)
data_1 <- matrix(rnorm(12),nrow=3,ncol=4,byrow=T)
data_2 <- matrix(rnorm(16),nrow=4,ncol=4,byrow=F)- and combine them.
data_new <- rbind(data_1,data_2)
data_new| -0.50219235 | 0.1315312 | -0.07891709 | 0.88678481 |
| 0.11697127 | 0.3186301 | -0.58179068 | 0.71453271 |
| -0.82525943 | -0.3598621 | 0.08988614 | 0.09627446 |
| -0.20163395 | -0.3888542 | -0.43808998 | -0.81437912 |
| 0.73984050 | 0.5108563 | 0.76406062 | -0.43845057 |
| 0.12337950 | -0.9138142 | 0.26196129 | -0.72022155 |
| -0.02931671 | 2.3102968 | 0.77340460 | 0.23094453 |
Indexing Matrices
set.seed(100) # to ensure the numbers are same for each of you
m <- matrix(rnorm(12), nrow = 3, ncol = 4, byrow = F)
m| -0.50219235 | 0.8867848 | -0.5817907 | -0.35986213 |
| 0.13153117 | 0.1169713 | 0.7145327 | 0.08988614 |
| -0.07891709 | 0.3186301 | -0.8252594 | 0.09627446 |
- Extract the first row.
m[1,]- -0.502192350531457
- 0.886784809417845
- -0.58179068471591
- -0.359862131395465
- Extract the second column.
m[,2]- 0.886784809417845
- 0.116971270510841
- 0.318630087617032
- Extract all the rows except the first row.
m[-1,]| 0.13153117 | 0.1169713 | 0.7145327 | 0.08988614 |
| -0.07891709 | 0.3186301 | -0.8252594 | 0.09627446 |
- Extract all the columns except the first and the third one.
m[, -c(1,3)]| 0.8867848 | -0.35986213 |
| 0.1169713 | 0.08988614 |
| 0.3186301 | 0.09627446 |
index_row <- 1:3
index_col <- c(1,3,4)- Extract the first, second and third row and first, third and fourth column.
m[index_row,index_col]| -0.50219235 | -0.5817907 | -0.35986213 |
| 0.13153117 | 0.7145327 | 0.08988614 |
| -0.07891709 | -0.8252594 | 0.09627446 |
- or alternatively you can use
m[1:3, c(1,3,4)]| -0.50219235 | -0.5817907 | -0.35986213 |
| 0.13153117 | 0.7145327 | 0.08988614 |
| -0.07891709 | -0.8252594 | 0.09627446 |
Data Frames
A data frame
- can be thought of as a data matrix or data set;
- is a list of vectors and/or factors of the same length;
- has a unique set of row names.
Data in the same position across columns come from the same experimental unit.
Can create data frames from pre-existing variables.
The main spec of data frame is the ability to keep variables with different forms.
Both numeric, string and logical variables can be reserved in a single dataframe unlike vectors and matrices.
Creata a vector called
mean_weight.
mean_weight <- c(179.3, 179.9, 180.5, 180.1, 180.3, 180.4)
mean_weight- 179.3
- 179.9
- 180.5
- 180.1
- 180.3
- 180.4
- Creata a vector called
Gender.
Gender <- c("M", "M", "F", "F", "M", "M")
Gender- 'M'
- 'M'
- 'F'
- 'F'
- 'M'
- 'M'
- Convert
Genderto a factor variable.
Gender <- factor(Gender,levels=c("M","F"))
Gender- M
- M
- F
- F
- M
- M
Levels:
- 'M'
- 'F'
- Combine both vectors into a dataframe.
d <- data.frame(mean_weight, Gender)
d| mean_weight | Gender |
|---|---|
| 179.3 | M |
| 179.9 | M |
| 180.5 | F |
| 180.1 | F |
| 180.3 | M |
| 180.4 | M |
- Note that the resulting variables have different data types.
mean_weightis numeric.Genderis factor.
- This wouldn’t be the case if we try to store them in a matrix as they can only store one type of variable.
Converting other Structures to Dataframes
You can also convert other data types to dataframes
- Converting a matrix to a data frame:
d <- cbind(mean_weight,Gender)
d| mean_weight | Gender |
|---|---|
| 179.3 | 1 |
| 179.9 | 1 |
| 180.5 | 2 |
| 180.1 | 2 |
| 180.3 | 1 |
| 180.4 | 1 |
- We created a matrix from
mean_weightandGender.Genderis automatically converted to a numerical variable as variables in the matrices should be in the same data type.
d <- as.data.frame(d)
d| mean_weight | Gender |
|---|---|
| 179.3 | 1 |
| 179.9 | 1 |
| 180.5 | 2 |
| 180.1 | 2 |
| 180.3 | 1 |
| 180.4 | 1 |
- Even if we convert the matrix to a dataframe the categorical names of the
Genderis gone.
Accesssing elements in a dataframe
- There are a lot of different way to access rows and columns in a dataframe.
- You can either use single bracket
[ ], double bracket[[ ]]or$sign. - Investigate the following code snippets to understand how
Rbehaves.
d$mean_weight # output in vector format- 179.3
- 179.9
- 180.5
- 180.1
- 180.3
- 180.4
d[["mean_weight"]] # output in vector format- 179.3
- 179.9
- 180.5
- 180.1
- 180.3
- 180.4
d[,1] # output in vector format- 179.3
- 179.9
- 180.5
- 180.1
- 180.3
- 180.4
d[,"mean_weight"] # output in vector format- 179.3
- 179.9
- 180.5
- 180.1
- 180.3
- 180.4
d["mean_weight"] # output in dataframe format| mean_weight |
|---|
| 179.3 |
| 179.9 |
| 180.5 |
| 180.1 |
| 180.3 |
| 180.4 |
d[1] # output in dataframe format| mean_weight |
|---|
| 179.3 |
| 179.9 |
| 180.5 |
| 180.1 |
| 180.3 |
| 180.4 |
- You can access a subset of rows by indexing the data frame
d[c(1,4,5),] # Shows 1., 4. and 5. rows of the dataframe| mean_weight | Gender | |
|---|---|---|
| 1 | 179.3 | 1 |
| 4 | 180.1 | 2 |
| 5 | 180.3 | 1 |
- It is suggested that you use
drop=FALSEwhen indexing (to sustain the data frame type).
d[1:3,"mean_weight"]- 179.3
- 179.9
- 180.5
d[1:3,"mean_weight",drop=FALSE]| mean_weight |
|---|
| 179.3 |
| 179.9 |
| 180.5 |
Creating a new variable in a dataframe
d| mean_weight | Gender |
|---|---|
| 179.3 | 1 |
| 179.9 | 1 |
| 180.5 | 2 |
| 180.1 | 2 |
| 180.3 | 1 |
| 180.4 | 1 |
d$color <- NA
d| mean_weight | Gender | color |
|---|---|---|
| 179.3 | 1 | NA |
| 179.9 | 1 | NA |
| 180.5 | 2 | NA |
| 180.1 | 2 | NA |
| 180.3 | 1 | NA |
| 180.4 | 1 | NA |
d$weight_two_times <- d$mean_weight*2
d| mean_weight | Gender | color | weight_two_times |
|---|---|---|---|
| 179.3 | 1 | NA | 358.6 |
| 179.9 | 1 | NA | 359.8 |
| 180.5 | 2 | NA | 361.0 |
| 180.1 | 2 | NA | 360.2 |
| 180.3 | 1 | NA | 360.6 |
| 180.4 | 1 | NA | 360.8 |
Importing Data
- The most popular functions for reading data sets
read.table()function is used mainly for reading data from formatted text files.read.csv()function is used mainly for reading data from files with csv format (“Comma Separated Values”format)read_excel()function is used to read data directly from an excel file. It requires the external packagereadxl.
Datasets
You can download the datasets used in this lecture from the lecturers AVESIS page.
Pima Data Set
- Indian females of Pima heritage (Native americans living in an area consisting of what is now central and southern Arizona)
- Columns (or Variables) of the Pima data set:
- NTP: number of times pregnant
- PGC: Plasma glucose concentration a 2 hours in an oral glucose tolerance test
- DBP: Diastolic blood pressure (mm Hg)
- TSFT: Triceps skin fold thickness (mm)
- SI: 2-Hour serum insulin (mu U/ml)
- BMI: Body mass index (weight in kg/(height in meter square))
- Diabetes pedigree function:
- Age: Age (years)
- Diabetes: f0,1g value 1 is interpreted as “tested positive for diabetes”
- First, you have to check whether the working directory of R and the location of file matches.
getwd()- The location of the working directory can be changed with
setwd()function. - Alternatively you can change the working directory to the where the Rcode is located with RStudio by using:
Session->Set Working Directory->To Source File Location
Working on the Posit Cloud
- If you are working on the Posit Cloud you don’t have to change your working directory since you are on the Cloud.
- But you have to upload your datasets to the cloud so that RStudio can locate and use it.
- On the left below pane, click the button Upload in Posit Cloud then a sub menu named Upload Files will emerge.
- Click Select Files from the menu
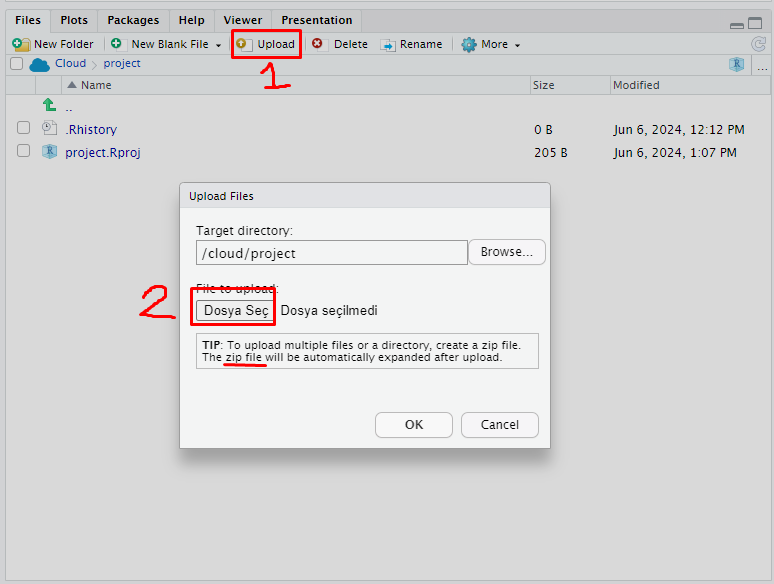
- From the file upload menu select the data files you wish to upload. If you intend to upload multiple files create a zip file from the datasets.
- Posit Cloud will automatically extract it once it uploads the zip file to the cloud.
- For example, week1_dataset.zip contains five different data formats for the pima dataset. When it is uploaded all five data files will be extracted under the folder week1_dataset.
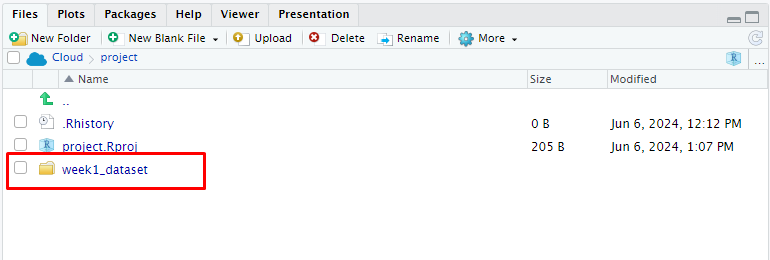
- Now you can use the following importing functions inside the Posit Cloud to call dataset into R.
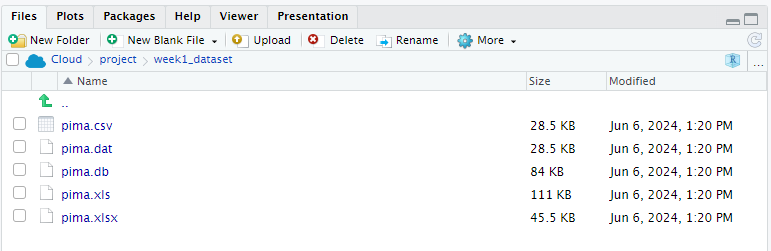
Importing Data from Text Files
- Now let’s check our working directory once more.
getwd()- Now if I want to import any data by using a function with R, I have to either:
- Provide the full location of the dataset inside the function such as:
C:/Users/erhan/Documents/FEF1002/pima.dat - or since my working directory is the folder
FEF1002, if I put mypima.datdata inside theFEF1002folder, it would be sufficient for me to providepima.datas the location.
- Provide the full location of the dataset inside the function such as:
- So in practice both
pima_data <- read.table("pima.dat", header = TRUE, sep = " ")and
You can use the head() function to see if everything is imported okay.
head(pima_data)| NTP | PGC | DBP | TSFT | SI | BMI | DPF | Age | Diabetes |
|---|---|---|---|---|---|---|---|---|
| 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | positive |
| 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | negative |
| 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | positive |
| 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | negative |
| 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | positive |
| 5 | 116 | 74 | 0 | 0 | 25.6 | 0.201 | 30 | negative |
- You can use the
str()function to see the structure of the dataset.
str(pima_data)
pima_data <- read.table("C:/Users/erhan/Documents/FEF1002/pima.dat",
header = TRUE, sep = " ")will work and import the data. * Remember to change C:/Users/erhan/Documents/FEF1002/pima.dat to where the pima.dat is actually located. * You can use both approach for the following data importing processes.
head(pima_data)| NTP | PGC | DBP | TSFT | SI | BMI | DPF | Age | Diabetes |
|---|---|---|---|---|---|---|---|---|
| 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | positive |
| 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | negative |
| 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | positive |
| 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | negative |
| 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | positive |
| 5 | 116 | 74 | 0 | 0 | 25.6 | 0.201 | 30 | negative |
str(pima_data)'data.frame': 768 obs. of 9 variables:
$ NTP : int 6 1 8 1 0 5 3 10 2 8 ...
$ PGC : int 148 85 183 89 137 116 78 115 197 125 ...
$ DBP : int 72 66 64 66 40 74 50 0 70 96 ...
$ TSFT : int 35 29 0 23 35 0 32 0 45 0 ...
$ SI : int 0 0 0 94 168 0 88 0 543 0 ...
$ BMI : num 33.6 26.6 23.3 28.1 43.1 25.6 31 35.3 30.5 0 ...
$ DPF : num 0.627 0.351 0.672 0.167 2.288 ...
$ Age : int 50 31 32 21 33 30 26 29 53 54 ...
$ Diabetes: Factor w/ 2 levels "negative","positive": 2 1 2 1 2 1 2 1 2 2 ...- Here, the argument
header = TRUEis used to denote that the variable names are given at the first line of the data. - The argument
sep = " "is used to denote how the variables are separated from each other. For this datasetspaceis used to separate variables.
Importing Data From CSV Files
pima_csv <- read.csv("pima.csv", header = TRUE, sep = ",")head(pima_csv)| NTP | PGC | DBP | TSFT | SI | BMI | DPF | Age | Diabetes |
|---|---|---|---|---|---|---|---|---|
| 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | positive |
| 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | negative |
| 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | positive |
| 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | negative |
| 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | positive |
| 5 | 116 | 74 | 0 | 0 | 25.6 | 0.201 | 30 | negative |
str(pima_csv)'data.frame': 768 obs. of 9 variables:
$ NTP : int 6 1 8 1 0 5 3 10 2 8 ...
$ PGC : int 148 85 183 89 137 116 78 115 197 125 ...
$ DBP : int 72 66 64 66 40 74 50 0 70 96 ...
$ TSFT : int 35 29 0 23 35 0 32 0 45 0 ...
$ SI : int 0 0 0 94 168 0 88 0 543 0 ...
$ BMI : num 33.6 26.6 23.3 28.1 43.1 25.6 31 35.3 30.5 0 ...
$ DPF : num 0.627 0.351 0.672 0.167 2.288 ...
$ Age : int 50 31 32 21 33 30 26 29 53 54 ...
$ Diabetes: Factor w/ 2 levels "negative","positive": 2 1 2 1 2 1 2 1 2 2 ...- From the code we can understand that column names are included in the data (
header = TRUE) and variables are separated withcomma(sep = ",")
Importing Data From Excel Files
- Remember that you can always use an external package to complete a different task
- Suppose we want to import directly from an excel file with
.xlsor.xlsxformat - We will use the
readxlpackage.
library(readxl) # Remember youj should use install.packages('readxl') if you didn't install it before
pima_xls <- read_excel("pima.xls", sheet = 'pima')head(pima_xls)| NTP | PGC | DBP | TSFT | SI | BMI | DPF | Age | Diabetes |
|---|---|---|---|---|---|---|---|---|
| 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | positive |
| 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | negative |
| 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | positive |
| 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | negative |
| 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | positive |
| 5 | 116 | 74 | 0 | 0 | 25.6 | 0.201 | 30 | negative |
str(pima_xls)Classes 'tbl_df', 'tbl' and 'data.frame': 768 obs. of 9 variables:
$ NTP : num 6 1 8 1 0 5 3 10 2 8 ...
$ PGC : num 148 85 183 89 137 116 78 115 197 125 ...
$ DBP : num 72 66 64 66 40 74 50 0 70 96 ...
$ TSFT : num 35 29 0 23 35 0 32 0 45 0 ...
$ SI : num 0 0 0 94 168 0 88 0 543 0 ...
$ BMI : num 33.6 26.6 23.3 28.1 43.1 25.6 31 35.3 30.5 0 ...
$ DPF : num 0.627 0.351 0.672 0.167 2.288 ...
$ Age : num 50 31 32 21 33 30 26 29 53 54 ...
$ Diabetes: chr "positive" "negative" "positive" "negative" ...pima_xlsx <- read_excel("pima.xlsx", sheet = 'pima')head(pima_xlsx)| NTP | PGC | DBP | TSFT | SI | BMI | DPF | Age | Diabetes |
|---|---|---|---|---|---|---|---|---|
| 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | positive |
| 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | negative |
| 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | positive |
| 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | negative |
| 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | positive |
| 5 | 116 | 74 | 0 | 0 | 25.6 | 0.201 | 30 | negative |
str(pima_xlsx)Classes 'tbl_df', 'tbl' and 'data.frame': 768 obs. of 9 variables:
$ NTP : num 6 1 8 1 0 5 3 10 2 8 ...
$ PGC : num 148 85 183 89 137 116 78 115 197 125 ...
$ DBP : num 72 66 64 66 40 74 50 0 70 96 ...
$ TSFT : num 35 29 0 23 35 0 32 0 45 0 ...
$ SI : num 0 0 0 94 168 0 88 0 543 0 ...
$ BMI : num 33.6 26.6 23.3 28.1 43.1 25.6 31 35.3 30.5 0 ...
$ DPF : num 0.627 0.351 0.672 0.167 2.288 ...
$ Age : num 50 31 32 21 33 30 26 29 53 54 ...
$ Diabetes: chr "positive" "negative" "positive" "negative" ...- We should define the sheet name of the data with the argument
sheetinside theread_excel()function.